Actor Partner Interdependence Model With Multilevel Analysis
Every time I teach multilevel modeling (MLM) at USC, I have students interested in running the actor partner independence model (APIM) using dyadic model. While such models are easier to fit with structural equation modeling, it can also be fit using MLM. In this post I provide a software tutorial for fitting such a model using MLM, and contrast to results to SEM.
Note that this is not a conceptual introduction to APIM. Please check out this chapter and this paper.
There is also this great tutorial on using the nlme package, which uses the dummy variable trick to allow a univariate MLM to handle multivariate analyses. The current blog post is similar but with a different package.
See also https://hydra.smith.edu/~rgarcia/workshop/Day_1-Actor-Partner_Interdependence_Model.html with the same example using nlme::gls().
Load Packages
library(tidyverse)
library(psych)
library(glmmTMB)Data
The data set comes from https://github.com/RandiLGarcia/dyadr/tree/master/data, which has 148 married couples. You can see the codebook at https://randilgarcia.github.io/week-dyad-workshop/Acitelli%20Codebook.pdf
pair_data <- here::here("data_files", "acipair.RData")
# Download data if not already exists in local folder
if (!file.exists(pair_data)) {
download.file("https://github.com/RandiLGarcia/dyadr/raw/master/data/acipair.RData",
pair_data)
}
load(pair_data)
# Show data
head(acipair)## CoupleID Yearsmar Partnum Gender_A SelfPos_A OtherPos_A Satisfaction_A
## 1 3 8.202667 1 Wife 4.8 4.6 4.000000
## 2 3 8.202667 2 Husband 3.8 4.0 3.666667
## 3 10 10.452667 1 Wife 4.6 3.8 3.166667
## 4 10 10.452667 2 Husband 4.2 4.0 3.666667
## 5 11 -8.297333 1 Wife 5.0 4.4 3.833333
## 6 11 -8.297333 2 Husband 4.2 4.8 3.833333
## Tension_A SimHob_A Gender_P SelfPos_P OtherPos_P Satisfaction_P Tension_P
## 1 1.5 0 MALE 3.8 4.0 3.666667 2.5
## 2 2.5 1 FEMALE 4.8 4.6 4.000000 1.5
## 3 4.0 0 MALE 4.2 4.0 3.666667 2.0
## 4 2.0 0 FEMALE 4.6 3.8 3.166667 4.0
## 5 2.5 0 MALE 4.2 4.8 3.833333 2.5
## 6 2.5 0 FEMALE 5.0 4.4 3.833333 2.5
## SimHob_P Gender COtherPos_A COtherPos_P CTension_A
## 1 1 Wife 0.3365 -0.2635 -0.9307
## 2 0 Husband -0.2635 0.3365 0.0693
## 3 0 Wife -0.4635 -0.2635 1.5693
## 4 0 Husband -0.2635 -0.4635 -0.4307
## 5 0 Wife 0.1365 0.5365 0.0693
## 6 0 Husband 0.5365 0.1365 0.0693
## CTension_P
## 1 0.0693
## 2 -0.9307
## 3 -0.4307
## 4 1.5693
## 5 0.0693
## 6 0.0693Hypothetical Research Question
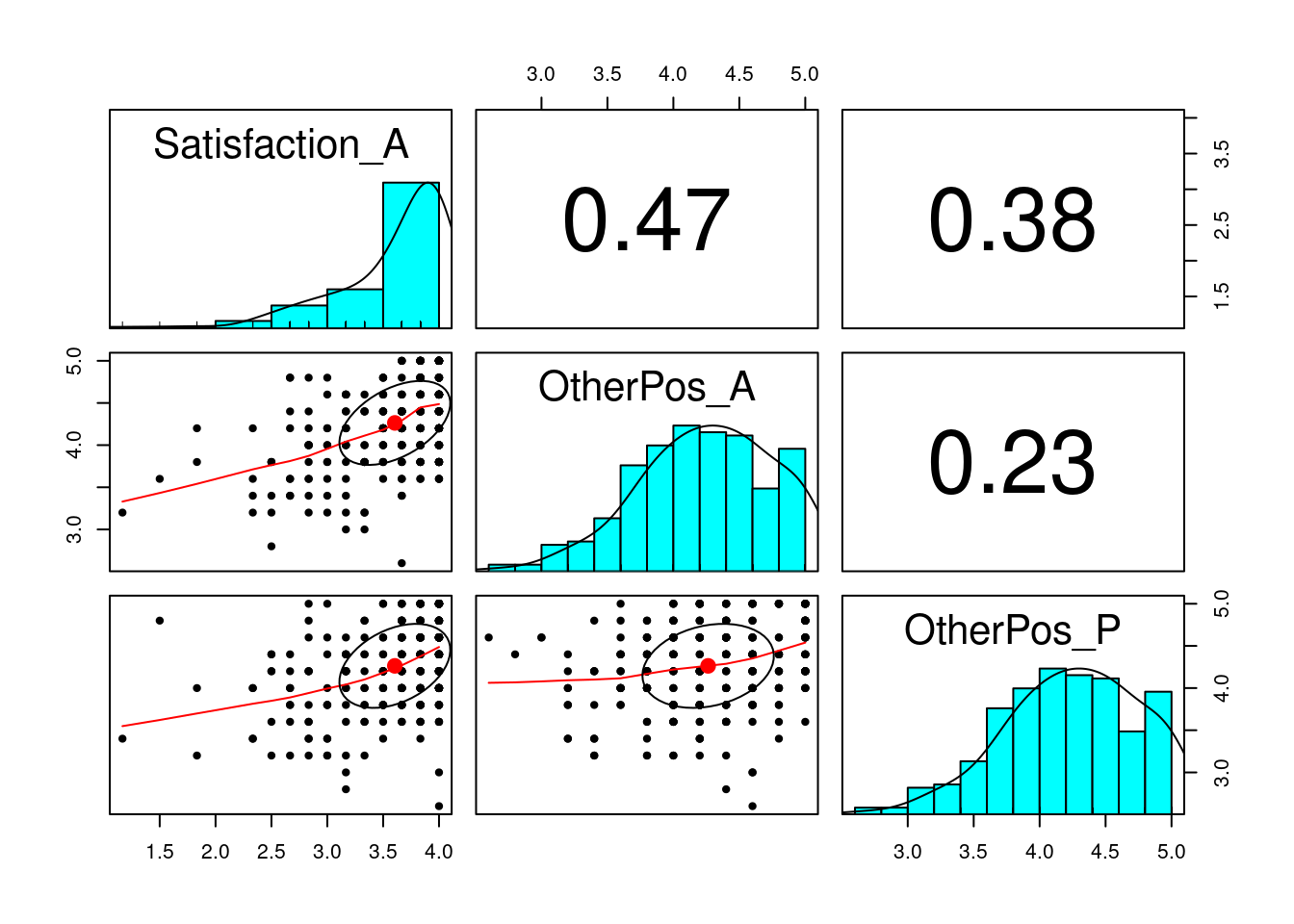
Here we have a hypothetical research question of how a person views their partners (OtherPos_A) and how their partner views them (OtherPos_P) predict their marriage satisfaction (Satisfaction_A).
# Overall
pairs.panels(
subset(acipair, select = c("Satisfaction_A", "OtherPos_A", "OtherPos_P"))
)
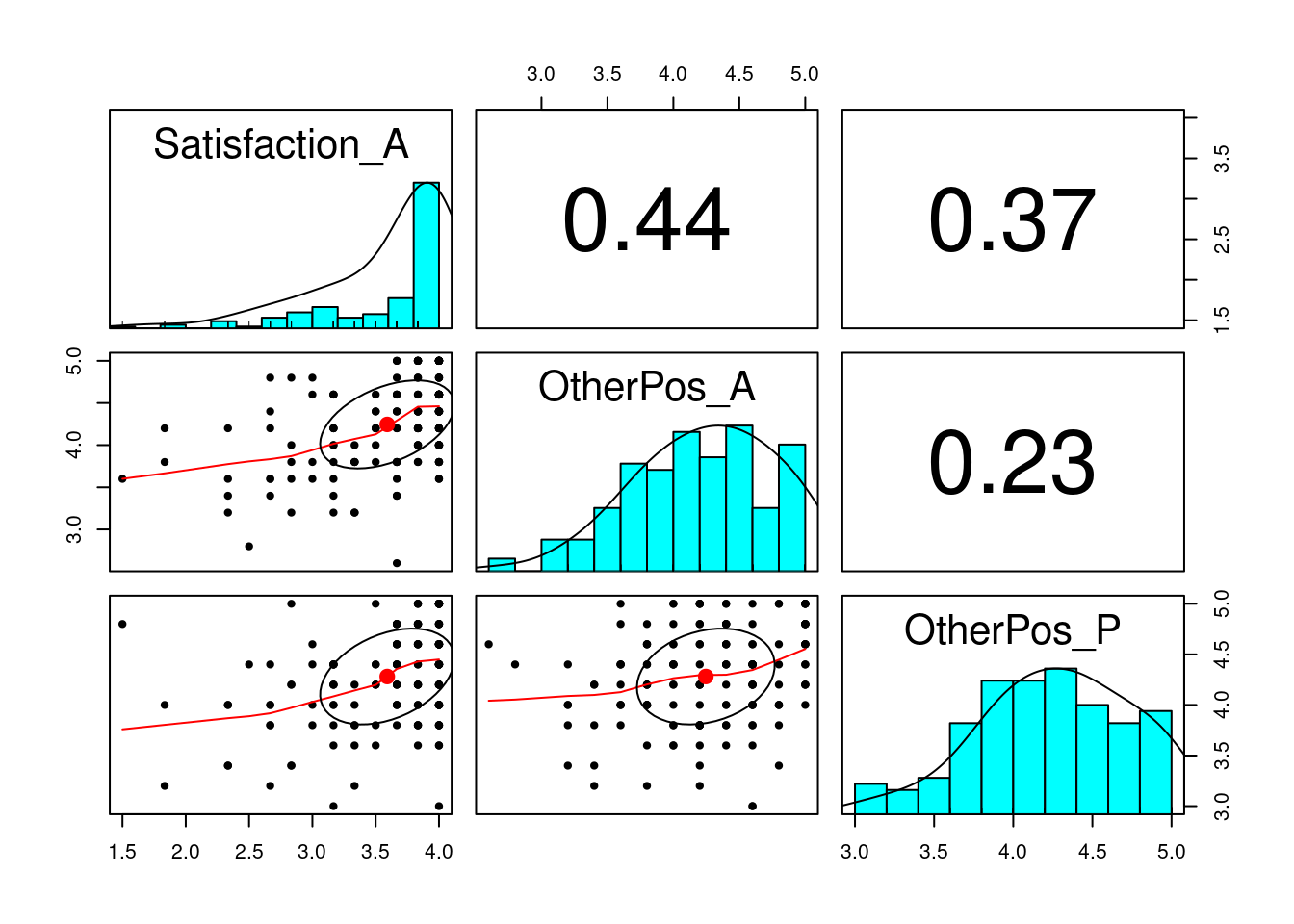
# Wife
pairs.panels(
subset(acipair, subset = Gender_A == "Wife",
select = c("Satisfaction_A", "OtherPos_A", "OtherPos_P"))
)
# Husband
pairs.panels(
subset(acipair, subset = Gender_A == "Husband",
select = c("Satisfaction_A", "OtherPos_A", "OtherPos_P"))
)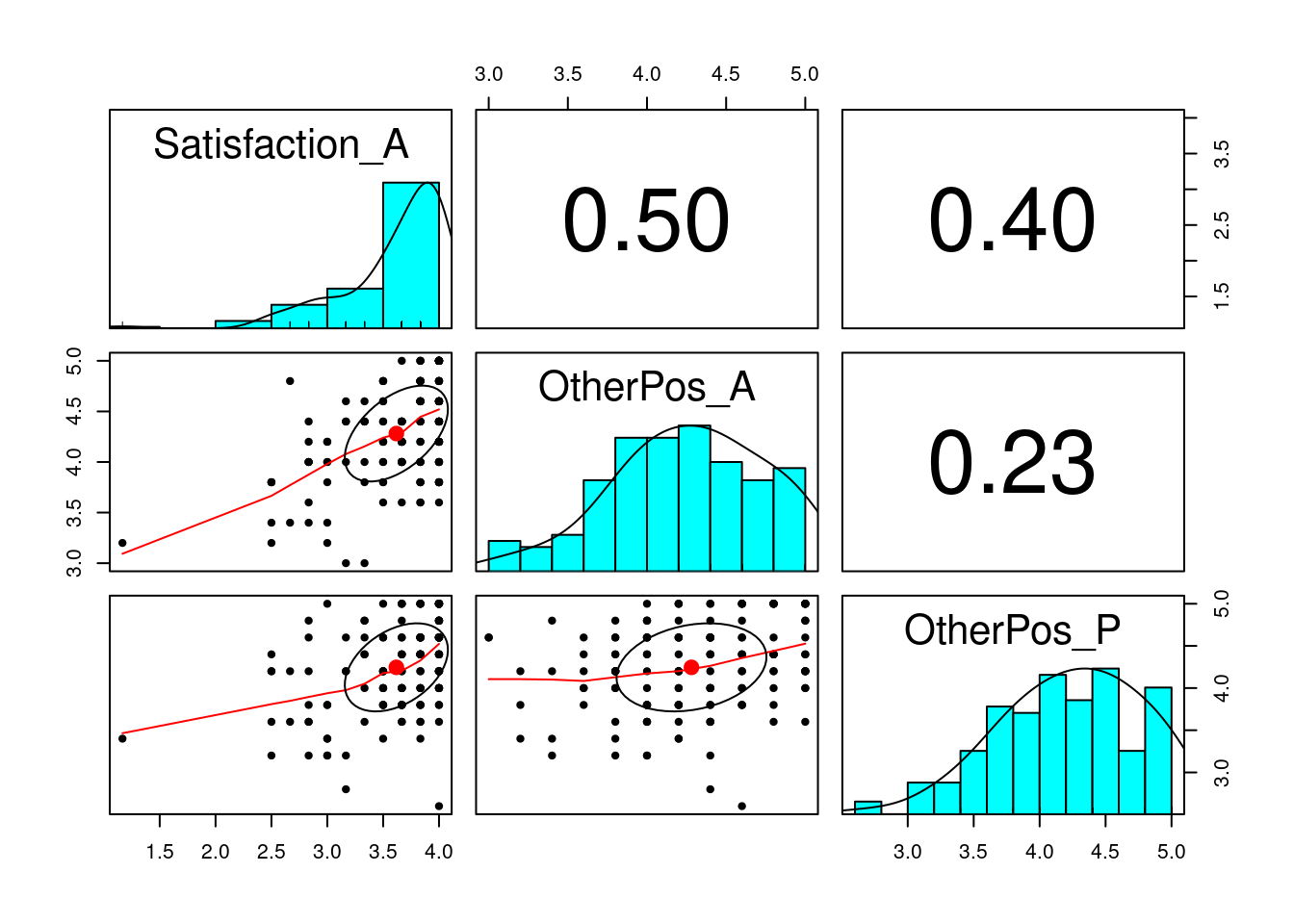
# Note the ceiling effect of satisfactionIn this example, there are two major paths of interest:
- Actor effect:
OtherPos_AtoSatisfaction_A - Partner effect:
OtherPos_PtoSatisfaction_A
In addition, for distinguishable dyads, like husband and wives, we want the gender interactions as well, resulting in four paths of interest (actor/partner effects for husbands/wives).
Distinguishable Dyads
Level 1:
\[ \begin{aligned} \text{Satisfaction}_{ij} & = \beta_{0j} + \beta_{1j} \text{View Partner}_{ij} + \beta_{2j} \text{Partner View}_{ij} + \beta_{3j} \text{Gender}_{ij} \\ & \quad + \beta_{4j} \text{View Partner}_{ij} \times \text{Gender}_{ij} \\ & \quad + \beta_{5j} \text{Partner View}_{ij} \times \text{Gender}_{ij} + e_{ij} \\ e_{ij} & \sim N(0, \sigma^2_{ij}) \\ \log(\sigma^2_{ij}) & = \beta^{s}_{0j} + \beta^{s}_{1j} \text{Gender}_{ij} \end{aligned} \]
Level 2:
\[ \begin{aligned} \beta_{0j} & = \gamma_{00} + u_{0j} \\ \beta_{1j} & = \gamma_{10} \\ \beta_{2j} & = \gamma_{20} \\ \beta_{3j} & = \gamma_{30} \\ \beta_{4j} & = \gamma_{40} \\ \beta_{5j} & = \gamma_{50} \\ \beta^{s}_{0j} & = \gamma^{s}_{00} \\ \beta^{s}_{1j} & = \gamma^{s}_{10} \\ u_{0j} | \text{Wife} & \sim N(0, \tau^2_{0 | H}) \\ u_{1j} | \text{Husband} & \sim N(0, \tau^2_{0 | W}) \end{aligned} \] Note that the variance components are allowed to be different across genders.
m_apim <- glmmTMB(
Satisfaction_A ~ (OtherPos_A + OtherPos_P) * Gender_A +
us(0 + Gender_A | CoupleID),
dispformula = ~ Gender_A,
data = acipair,
REML = FALSE
)
summary(m_apim)## Family: gaussian ( identity )
## Formula:
## Satisfaction_A ~ (OtherPos_A + OtherPos_P) * Gender_A + us(0 +
## Gender_A | CoupleID)
## Dispersion: ~Gender_A
## Data: acipair
##
## AIC BIC logLik deviance df.resid
## 297.6 338.2 -137.8 275.6 285
##
## Random effects:
##
## Conditional model:
## Groups Name Variance Std.Dev. Corr
## CoupleID Gender_AWife 0.08734 0.2955
## Gender_AHusband 0.07716 0.2778 0.98
## Residual NA NA
## Number of obs: 296, groups: CoupleID, 148
##
## Conditional model:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) 0.61125 0.40861 1.496 0.135
## OtherPos_A 0.37770 0.07317 5.162 2.44e-07 ***
## OtherPos_P 0.32148 0.08069 3.984 6.78e-05 ***
## Gender_AHusband 0.07921 0.38779 0.204 0.838
## OtherPos_A:Gender_AHusband 0.04669 0.10458 0.446 0.655
## OtherPos_P:Gender_AHusband -0.05983 0.10628 -0.563 0.573
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Dispersion model:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -2.1549 262.4017 -0.008 0.993
## Gender_AHusband -0.6085 680.1472 -0.001 0.999The model results show:
- Actor effect for Wife: 0.37 (SE = 0.07)
- Partner effect for Wife: 0.32 (SE = 0.08)
- Difference of actor effect (Husband - Wife): 0.05 (SE = 0.10)
- Difference of partner effect (Husband - Wife): -0.06 (SE = 0.11)
- Gender difference (Husband - Wife) when Partner View is 0: 0.08 (SE = 0.39)
We can also estimate the actor and partner effects for both Husband and Wife by suppressing the intercept:
m_apim2 <- glmmTMB(
Satisfaction_A ~ 0 + Gender_A + (OtherPos_A + OtherPos_P):Gender_A +
us(0 + Gender_A | CoupleID),
dispformula = ~ Gender_A,
data = acipair,
REML = FALSE,
# The default optimizer did not converge; try optim
control = glmmTMBControl(
optimizer = optim,
optArgs = list(method = "BFGS")
)
)
summary(m_apim2)## Family: gaussian ( identity )
## Formula:
## Satisfaction_A ~ 0 + Gender_A + (OtherPos_A + OtherPos_P):Gender_A +
## us(0 + Gender_A | CoupleID)
## Dispersion: ~Gender_A
## Data: acipair
##
## AIC BIC logLik deviance df.resid
## 297.6 338.2 -137.8 275.6 285
##
## Random effects:
##
## Conditional model:
## Groups Name Variance Std.Dev. Corr
## CoupleID Gender_AWife 0.11075 0.3328
## Gender_AHusband 0.05813 0.2411 1.00
## Residual NA NA
## Number of obs: 296, groups: CoupleID, 148
##
## Conditional model:
## Estimate Std. Error z value Pr(>|z|)
## Gender_AWife 0.61125 0.40861 1.496 0.1347
## Gender_AHusband 0.69046 0.33941 2.034 0.0419 *
## Gender_AWife:OtherPos_A 0.37770 0.07317 5.162 2.44e-07 ***
## Gender_AHusband:OtherPos_A 0.42439 0.06703 6.332 2.43e-10 ***
## Gender_AWife:OtherPos_P 0.32148 0.08069 3.984 6.78e-05 ***
## Gender_AHusband:OtherPos_P 0.26165 0.06078 4.305 1.67e-05 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Dispersion model:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -2.3805 94.5039 -0.025 0.980
## Gender_AHusband -0.1192 150.4151 -0.001 0.999Indistinguishable Dyads
As the interaction terms were not significant, one may want to remove them. Similarly, the variance components did not look too different across genders, so we may make them equal as well:
m2 <- glmmTMB(
Satisfaction_A ~ Gender_A + OtherPos_A + OtherPos_P + (1 | CoupleID),
dispformula = ~ 1,
data = acipair,
REML = FALSE
)
summary(m2)## Family: gaussian ( identity )
## Formula:
## Satisfaction_A ~ Gender_A + OtherPos_A + OtherPos_P + (1 | CoupleID)
## Data: acipair
##
## AIC BIC logLik deviance df.resid
## 294.5 316.6 -141.2 282.5 290
##
## Random effects:
##
## Conditional model:
## Groups Name Variance Std.Dev.
## CoupleID (Intercept) 0.08047 0.2837
## Residual 0.09155 0.3026
## Number of obs: 296, groups: CoupleID, 148
##
## Dispersion estimate for gaussian family (sigma^2): 0.0915
##
## Conditional model:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) 0.65818 0.32074 2.052 0.0402 *
## Gender_AHusband 0.02315 0.03523 0.657 0.5111
## OtherPos_A 0.39935 0.04706 8.487 < 2e-16 ***
## OtherPos_P 0.28904 0.04706 6.143 8.12e-10 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Finally, we may also remove the main effect for Gender:
m3 <- glmmTMB(
Satisfaction_A ~ OtherPos_A + OtherPos_P + (1 | CoupleID),
dispformula = ~ 1,
data = acipair,
REML = FALSE
)
summary(m3)## Family: gaussian ( identity )
## Formula: Satisfaction_A ~ OtherPos_A + OtherPos_P + (1 | CoupleID)
## Data: acipair
##
## AIC BIC logLik deviance df.resid
## 292.9 311.3 -141.4 282.9 291
##
## Random effects:
##
## Conditional model:
## Groups Name Variance Std.Dev.
## CoupleID (Intercept) 0.08034 0.2834
## Residual 0.09181 0.3030
## Number of obs: 296, groups: CoupleID, 148
##
## Dispersion estimate for gaussian family (sigma^2): 0.0918
##
## Conditional model:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) 0.66975 0.32025 2.091 0.0365 *
## OtherPos_A 0.40042 0.04705 8.510 < 2e-16 ***
## OtherPos_P 0.28797 0.04705 6.120 9.34e-10 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Model Comparison
anova(m_apim, m2, m3)## Data: acipair
## Models:
## m3: Satisfaction_A ~ OtherPos_A + OtherPos_P + (1 | CoupleID), zi=~0, disp=~1
## m2: Satisfaction_A ~ Gender_A + OtherPos_A + OtherPos_P + (1 | CoupleID), zi=~0, disp=~1
## m_apim: Satisfaction_A ~ (OtherPos_A + OtherPos_P) * Gender_A + us(0 + , zi=~0, disp=~Gender_A
## m_apim: Gender_A | CoupleID), zi=~0, disp=~1
## Df AIC BIC logLik deviance Chisq Chi Df Pr(>Chisq)
## m3 5 292.88 311.34 -141.44 282.88
## m2 6 294.45 316.60 -141.23 282.45 0.4312 1 0.5114
## m_apim 11 297.61 338.20 -137.80 275.61 6.8458 5 0.2324There model with all paths equal is the best in terms of AIC and BIC, and the likelihood ratio tests were not significant.
Using SEM
Distinguishable Dyads
# Wide format
aciwide <- filter(acipair, Partnum == 1) # Actor = Wife; Partner = Husband
library(lavaan)## This is lavaan 0.6-11
## lavaan is FREE software! Please report any bugs.##
## Attaching package: 'lavaan'## The following object is masked from 'package:psych':
##
## cor2covapim_mod <- ' Satisfaction_A + Satisfaction_P ~ OtherPos_A + OtherPos_P '
apim_fit <- sem(apim_mod, data = aciwide)
summary(apim_fit)## lavaan 0.6-11 ended normally after 12 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 7
##
## Number of observations 148
##
## Model Test User Model:
##
## Test statistic 0.000
## Degrees of freedom 0
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Regressions:
## Estimate Std.Err z-value P(>|z|)
## Satisfaction_A ~
## OtherPos_A 0.378 0.073 5.162 0.000
## OtherPos_P 0.321 0.081 3.984 0.000
## Satisfaction_P ~
## OtherPos_A 0.262 0.061 4.305 0.000
## OtherPos_P 0.424 0.067 6.332 0.000
##
## Covariances:
## Estimate Std.Err z-value P(>|z|)
## .Satisfaction_A ~~
## .Satisfaction_P 0.080 0.015 5.221 0.000
##
## Variances:
## Estimate Std.Err z-value P(>|z|)
## .Satisfaction_A 0.203 0.024 8.602 0.000
## .Satisfaction_P 0.140 0.016 8.602 0.000library(semPlot)
library(semptools)
p1 <- semPaths(apim_fit, what = "est",
rotation = 2,
sizeMan = 15,
nCharNodes = 0,
edge.label.cex = 1.15,
label.cex = 1.25)my_label_list <- list(list(node = "Satisfaction_A", to = "Satisfaction (W)"),
list(node = "Satisfaction_P", to = "Satisfaction (H)"),
list(node = "OtherPos_A", to = "View\nPartner (W)"),
list(node = "OtherPos_P", to = "View\nPartner (H)"))
p2 <- change_node_label(p1, my_label_list)
p3 <- mark_se(p2, apim_fit, sep = "\n")
my_position_list <- c("Satisfaction_A ~ OtherPos_P" = .25,
"Satisfaction_P ~ OtherPos_A" = .25)
p4 <- set_edge_label_position(p3, my_position_list)
plot(p4)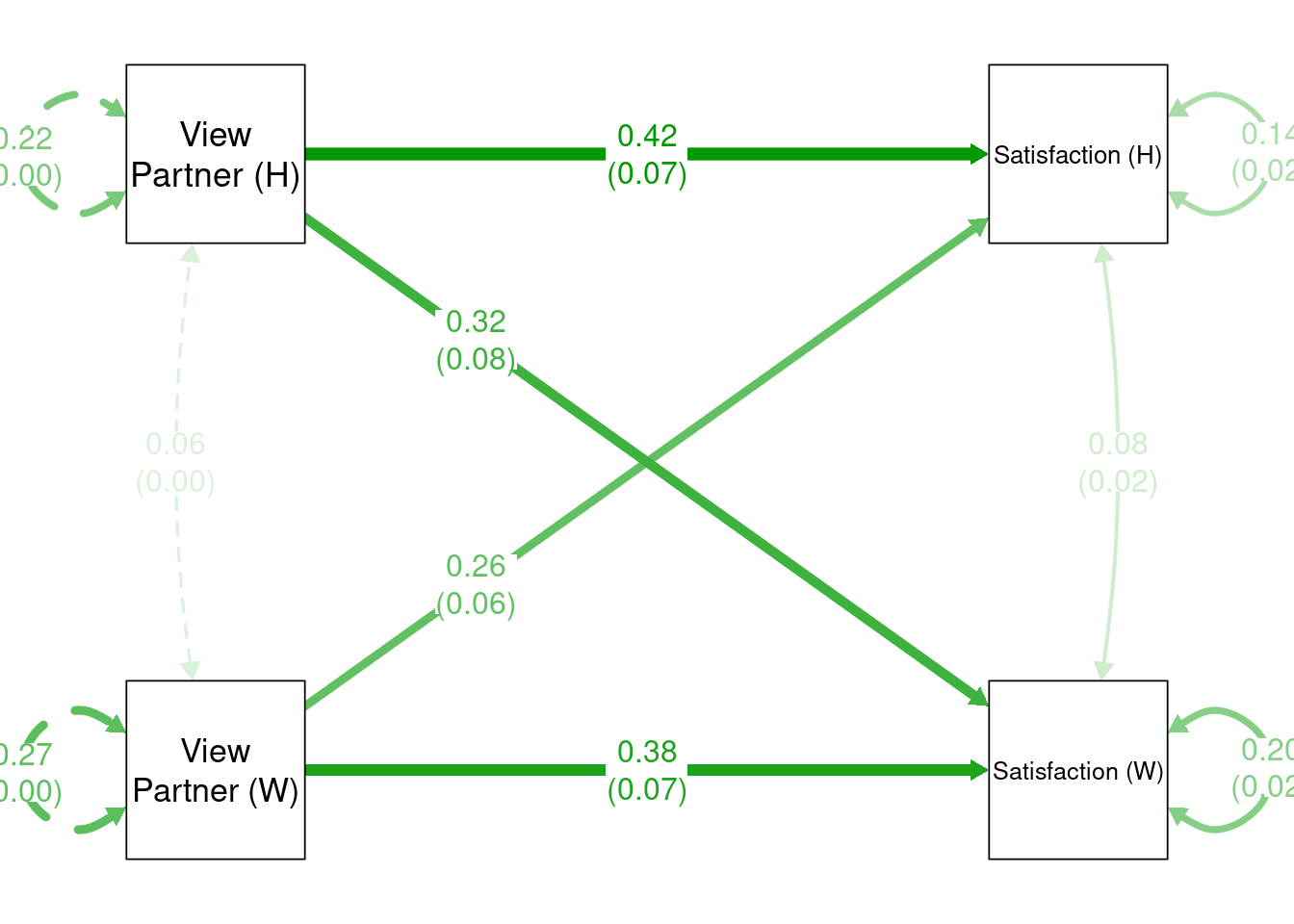
Indistinguishable Dyads
apim2_mod <- ' Satisfaction_A ~ a * OtherPos_A + p * OtherPos_P
Satisfaction_P ~ p * OtherPos_A + a * OtherPos_P
# Constrain the variances and means to be equal
Satisfaction_A ~~ v * Satisfaction_A
Satisfaction_P ~~ v * Satisfaction_P
Satisfaction_A ~ m * 1
Satisfaction_P ~ m * 1 '
apim2_fit <- sem(apim2_mod, data = aciwide)
summary(apim2_fit)## lavaan 0.6-11 ended normally after 22 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 9
## Number of equality constraints 4
##
## Number of observations 148
##
## Model Test User Model:
##
## Test statistic 7.277
## Degrees of freedom 4
## P-value (Chi-square) 0.122
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Regressions:
## Estimate Std.Err z-value P(>|z|)
## Satisfaction_A ~
## OtherPos_A (a) 0.400 0.047 8.510 0.000
## OtherPos_P (p) 0.288 0.047 6.120 0.000
## Satisfaction_P ~
## OtherPos_A (p) 0.288 0.047 6.120 0.000
## OtherPos_P (a) 0.400 0.047 8.510 0.000
##
## Covariances:
## Estimate Std.Err z-value P(>|z|)
## .Satisfaction_A ~~
## .Satisfaction_P 0.080 0.016 5.145 0.000
##
## Intercepts:
## Estimate Std.Err z-value P(>|z|)
## .Satsfctn_A (m) 0.670 0.320 2.091 0.036
## .Satsfctn_P (m) 0.670 0.320 2.091 0.036
##
## Variances:
## Estimate Std.Err z-value P(>|z|)
## .Satsfctn_A (v) 0.172 0.016 11.024 0.000
## .Satsfctn_P (v) 0.172 0.016 11.024 0.000p1 <- semPaths(apim2_fit, what = "est",
rotation = 2,
sizeMan = 15,
nCharNodes = 0,
edge.label.cex = 1.15,
label.cex = 1.25,
intercepts = FALSE)my_label_list <- list(list(node = "Satisfaction_A", to = "Satisfaction (W)"),
list(node = "Satisfaction_P", to = "Satisfaction (H)"),
list(node = "OtherPos_A", to = "View\nPartner (W)"),
list(node = "OtherPos_P", to = "View\nPartner (H)"))
p2 <- change_node_label(p1, my_label_list)
p3 <- mark_se(p2, apim2_fit, sep = "\n")
my_position_list <- c("Satisfaction_A ~ OtherPos_P" = .25,
"Satisfaction_P ~ OtherPos_A" = .25)
p4 <- set_edge_label_position(p3, my_position_list)
plot(p4)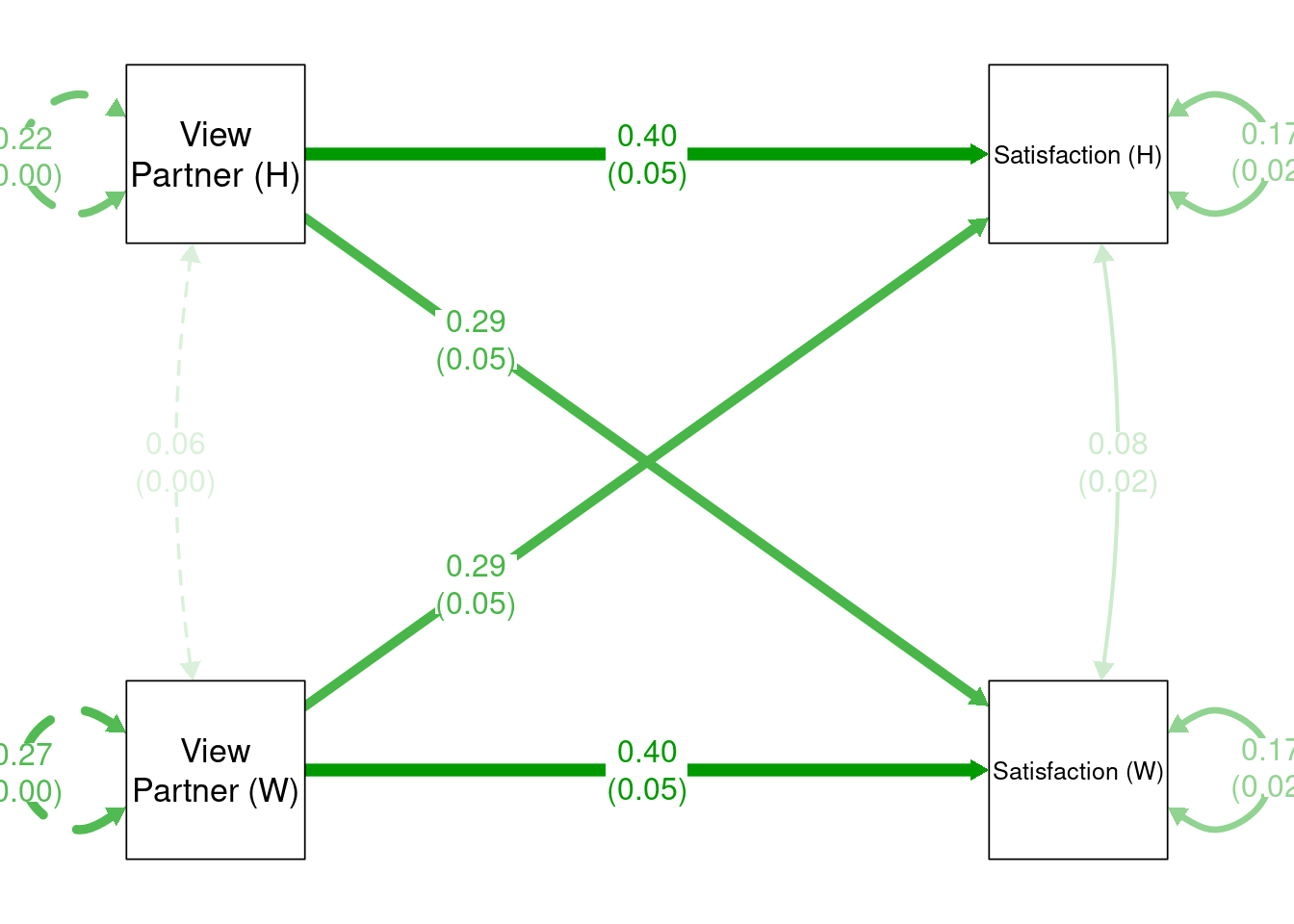
The results are basically identical.