Confidence Intervals for Multilevel R-Squared
Load Packages
library(lme4)## Loading required package: Matrixlibrary(MuMIn) # for computing multilevel R-squared
library(r2mlm) # another package for R-squared## Loading required package: nlme##
## Attaching package: 'nlme'## The following object is masked from 'package:lme4':
##
## lmList## Registered S3 method overwritten by 'parameters':
## method from
## format.parameters_distribution datawizardlibrary(bootmlm) # for multilevel bootstrapping
library(boot) # for bootstrap CIsAn Example Multilevel Model
fm1 <- lmer(Reaction ~ Days + (Days | Subject), sleepstudy)Nakagawa-Johnson-Schielzeth \(R^2\)
r.squaredGLMM(fm1)## Warning: 'r.squaredGLMM' now calculates a revised statistic. See the help page.## R2m R2c
## [1,] 0.2786511 0.7992199The marginal \(R^2\) considers the total variance accounted for due to the fixed effect associated with the predictors (Days in this example). See Nakagawa, Johnson, & Schielzeth (2017) for more information.
Right-Sterba \(R^2\)
More fine-grained partitioning, as described in Rights & Sterba (2019)
r2mlm(fm1)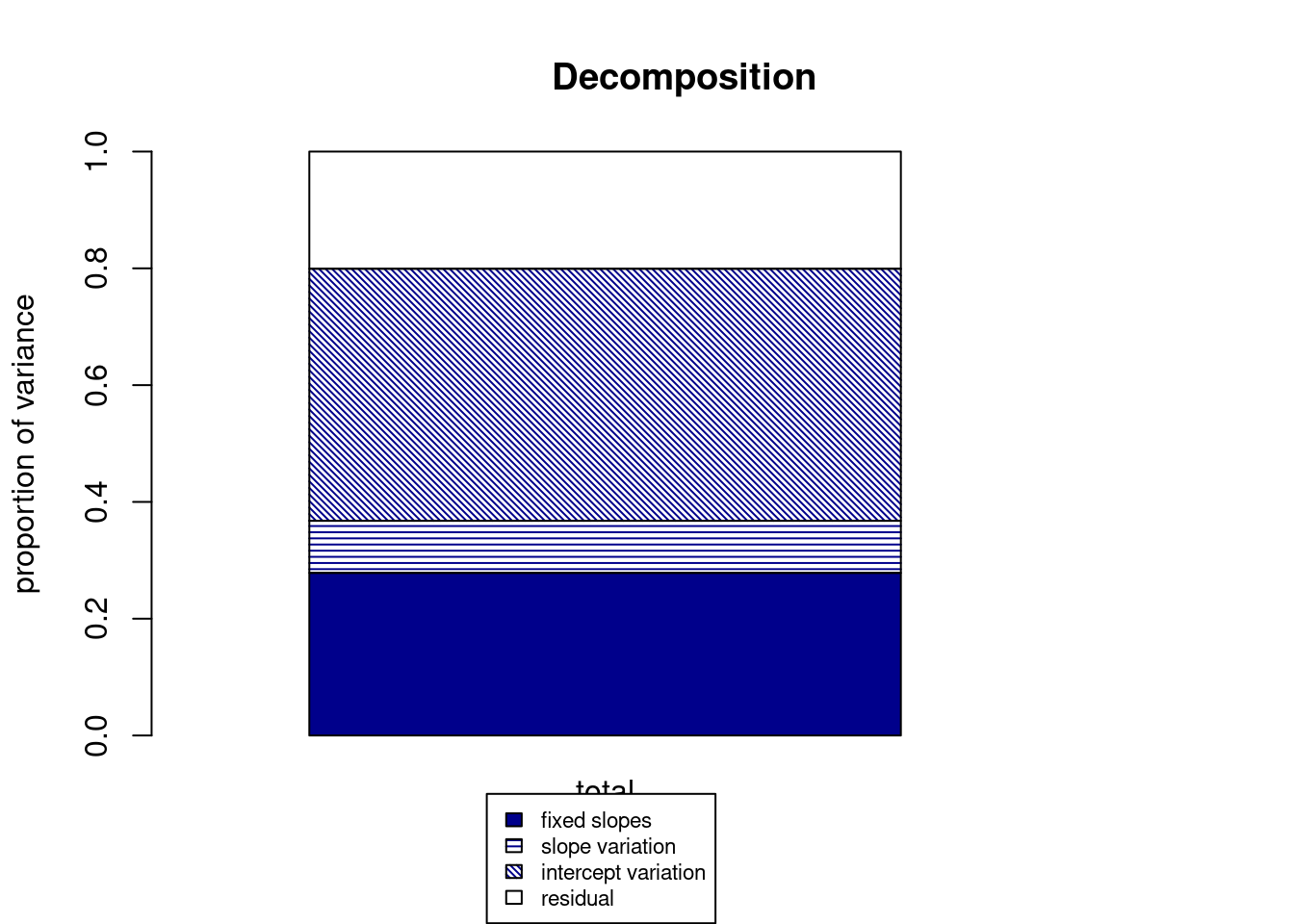
## $Decompositions
## total
## fixed 0.27851304
## slope variation 0.08915267
## mean variation 0.43165365
## sigma2 0.20068063
##
## $R2s
## total
## f 0.27851304
## v 0.08915267
## m 0.43165365
## fv 0.36766572
## fvm 0.79931937The fixed part is the same as the marginal \(R^2\).
Confidence Intervals for \(R^2\)
Neither MuMIn::r.squaredGLMM() nor r2mlm::r2mlm() provided confidence intervals (CIs) for the \(R^2\), but general guidelines for effect size reporting would suggest always reporting CIs for point estimates of effect size, just like for any point estimates in statistics. We can use multilevel bootstrapping to get CIs.
To do bootstrap, first defines an R function that gives the target \(R^2\) statistics. We can do it for the marginal \(R^2\):
marginal_r2 <- function(object) {
r.squaredGLMM(object)[[1]]
}
marginal_r2(fm1)## [1] 0.2786511Parametric Bootstrap
The lme4::bootMer() supports basic parametric multilevel bootstrapping
# This takes about 30 sec on my computer
boo01 <- bootMer(fm1, FUN = marginal_r2, nsim = 999)
boo01##
## PARAMETRIC BOOTSTRAP
##
##
## Call:
## bootMer(x = fm1, FUN = marginal_r2, nsim = 999)
##
##
## Bootstrap Statistics :
## original bias std. error
## t1* 0.2786511 0.005759548 0.07545455##
## 19 message(s): boundary (singular) fit: see help('isSingular')
## 9 warning(s): Model failed to converge with max|grad| = 0.00203576 (tol = 0.002, component 1) (and others)Here is the bootstrap distribution
plot(boo01)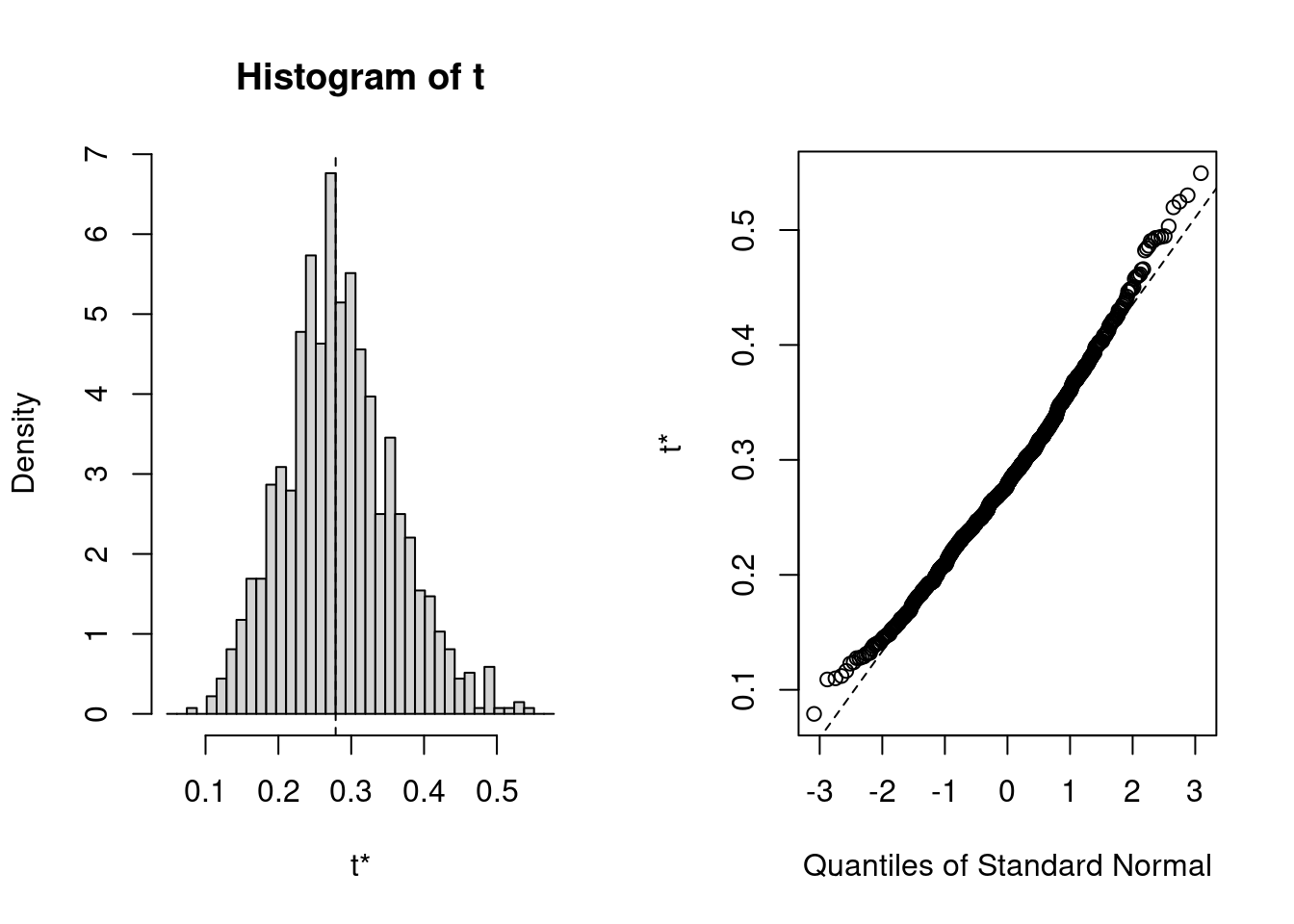
Bias-corrected estimate
The above shows that the sample estimate of \(R^2\) was upwardly biased. To correct for the bias, we can use the bootstrap bias-corrected estimate
2 * boo01$t0 - mean(boo01$t)## [1] 0.2728915Confidence intervals
You can get three types of bootstrap CIs ("norm", "basic", "perc") with bootMer:
boot::boot.ci(boo01, index = 1, type = c("norm", "basic", "perc"))## BOOTSTRAP CONFIDENCE INTERVAL CALCULATIONS
## Based on 999 bootstrap replicates
##
## CALL :
## boot::boot.ci(boot.out = boo01, type = c("norm", "basic", "perc"),
## index = 1)
##
## Intervals :
## Level Normal Basic Percentile
## 95% ( 0.1250, 0.4208 ) ( 0.1088, 0.4114 ) ( 0.1460, 0.4485 )
## Calculations and Intervals on Original ScaleResidual Bootstrap
The bootmlm::bootstrap_mer() implements the residual bootstrap, which is robust to non-normality.
# This takes about 30 sec on my computer
boo02 <- bootstrap_mer(fm1, FUN = marginal_r2, nsim = 999, type = "residual")## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')boo02##
## ORDINARY NONPARAMETRIC BOOTSTRAP
##
##
## Call:
## bootstrap_mer(x = fm1, FUN = marginal_r2, nsim = 999, type = "residual")
##
##
## Bootstrap Statistics :
## original bias std. error
## t1* 0.2786511 0.005397786 0.08418536In this example, the results are similar. The boostrap bias-corrected estimate of \(R^2\), and the three basic CIs, can similarly be computed as in parametric bootstrap.
Confidence Intervals
In addition to the three CIs previously discussed, which are first-order accurate, we can also obtain CIs that are second-order accurate: (a) bias-corrected and accelerated (BCa) CI and (b) studentized CI (also called the bootstrap-\(t\) CI). For (a), it requires the influence value of the \(R^2\) function, whereas for (b), it requires an estimate of the sampling variance of the \(R^2\) estimate.
Influence value
# Based on the group jackknife
inf_val <- bootmlm::empinf_mer(fm1, marginal_r2, index = 1)Sampling variance with the numerical delta method
This part is a bit more technical; skip this if you’re not interested in the studentized CI.
To obtain an approximate sampling variance of the \(R^2\), it would be easier to use the r2mlm::r2mlm_manual() function to compute \(R^2\). We first write a function that computes \(R^2\) using input of the fixed and random effects:
manual_r2 <- function(theta, data,
# The following are model-specific
wc = 2, bc = NULL, rc = 2,
cmc = FALSE) {
n_wc <- length(wc)
n_bc <- length(bc) + 1
dim_rc <- length(rc) + 1
n_rc <- dim_rc * (dim_rc + 1) / 2
gam_w <- theta[seq_len(n_wc)]
gam_b <- theta[n_wc + seq_len(n_bc)]
tau <- matrix(NA, nrow = dim_rc, ncol = dim_rc)
tau[lower.tri(tau, diag = TRUE)] <- theta[n_wc + n_bc + seq_len(n_rc)]
tau2 <- t(tau)
tau2[lower.tri(tau2)] <- tau[lower.tri(tau)]
s2 <- tail(theta, n = 1)
r2mlm_manual(data,
within_covs = wc,
between_covs = bc,
random_covs = 2,
gamma_w = gam_w,
gamma_b = gam_b,
Tau = tau2,
sigma2 = s2,
clustermeancentered = cmc,
bargraph = FALSE)$R2s[1, 1]
}
theta_fm1 <- c(fixef(fm1)[2], fixef(fm1)[1],
VarCorr(fm1)[[1]][c(1, 2, 4)], sigma(fm1)^2)
manual_r2(theta_fm1, data = fm1@frame)## [1] 0.278513Now computes the numerical gradient
grad_fm1 <- numDeriv::grad(manual_r2, x = theta_fm1, data = fm1@frame)We also need the asymptotic covariance matrix of the fixed and random effects
vcov_fixed <- vcov(fm1)
vcov_random <- vcov_vc(fm1, sd_cor = FALSE, print_names = FALSE)
vcov_fm1 <- bdiag(vcov_fixed, vcov_random)
# Need to re-arrange the first two columns
vcov_fm1 <- vcov_fm1[c(2, 1, 3:6), c(2, 1, 3:6)]Now apply the multivariate delta method
crossprod(grad_fm1, vcov_fm1) %*% grad_fm1## 1 x 1 Matrix of class "dgeMatrix"
## [,1]
## [1,] 0.005919918We now need a function that computes both \(R^2\) and the asymptotic sampling variance of it.
marginal_r2_with_var <- function(object,
wc = 2, bc = NULL, rc = 2) {
dim_rc <- length(rc) + 1
vc_mat <- matrix(seq_len(dim_rc^2), nrow = dim_rc, ncol = dim_rc)
vc_index <- vc_mat[lower.tri(vc_mat, diag = TRUE)]
theta_obj <- c(fixef(object)[wc], fixef(object)[c(1, bc)],
VarCorr(object)[[1]][vc_index], sigma(object)^2)
r2 <- manual_r2(theta_obj, data = object@frame)
grad_obj <- numDeriv::grad(manual_r2, x = theta_obj, data = object@frame)
# Need to re-arrange the order of the fixed effects
names_wc <- names(object@frame)[wc]
names_bc <- c("(Intercept)", names(object@frame)[bc])
vcov_fixed <- vcov(object)[c(names_wc, names_bc), c(names_wc, names_bc)]
vcov_random <- vcov_vc(object, sd_cor = FALSE, print_names = FALSE)
vcov_obj <- bdiag(vcov_fixed, vcov_random)
v_r2 <- crossprod(grad_obj, vcov_obj) %*% grad_obj
c(r2, as.numeric(v_r2))
}
marginal_r2_with_var(fm1)## [1] 0.278513044 0.005919918Five Bootstrap CIs
Now, we can do bootstrap again, using the new function that computes both the estimate and the asymptotic sampling variance
# This takes quite a bit longer due to the need to compute variances
boo03 <- bootstrap_mer(fm1, FUN = marginal_r2_with_var, nsim = 999,
type = "residual")## boundary (singular) fit: see help('isSingular')## Warning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
## Model failed to converge with max|grad| = 0.00828807 (tol = 0.002, component 1)## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')## Warning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
## Model failed to converge with max|grad| = 0.00967386 (tol = 0.002, component 1)## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')## Warning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
## Model failed to converge with max|grad| = 0.00453875 (tol = 0.002, component 1)## boundary (singular) fit: see help('isSingular')## Warning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
## Model failed to converge with max|grad| = 0.00280723 (tol = 0.002, component 1)## boundary (singular) fit: see help('isSingular')## Warning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
## Model failed to converge with max|grad| = 0.00764723 (tol = 0.002, component 1)## Warning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
## Model failed to converge with max|grad| = 0.0024541 (tol = 0.002, component 1)## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')## Warning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
## Model failed to converge with max|grad| = 0.0049756 (tol = 0.002, component 1)## Warning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
## Model failed to converge with max|grad| = 0.00228953 (tol = 0.002, component 1)## boundary (singular) fit: see help('isSingular')## Warning in sqrt(diag(m)): NaNs produced## Warning in sqrt(diag(m)): NaNs produced
## Warning in sqrt(diag(m)): NaNs produced
## Warning in sqrt(diag(m)): NaNs produced
## Warning in sqrt(diag(m)): NaNs produced
## Warning in sqrt(diag(m)): NaNs produced
## Warning in sqrt(diag(m)): NaNs produced
## Warning in sqrt(diag(m)): NaNs produced
## Warning in sqrt(diag(m)): NaNs produced
## Warning in sqrt(diag(m)): NaNs produced
## Warning in sqrt(diag(m)): NaNs produced
## Warning in sqrt(diag(m)): NaNs produced## boundary (singular) fit: see help('isSingular')
## boundary (singular) fit: see help('isSingular')boo03##
## ORDINARY NONPARAMETRIC BOOTSTRAP
##
##
## Call:
## bootstrap_mer(x = fm1, FUN = marginal_r2_with_var, nsim = 999,
## type = "residual")
##
##
## Bootstrap Statistics :
## original bias std. error
## t1* 0.278513044 0.0100127766 0.083704256
## t2* 0.005919918 -0.0002646922 0.001150678And then obtain five types of CIs
boot::boot.ci(boo03, L = inf_val)## BOOTSTRAP CONFIDENCE INTERVAL CALCULATIONS
## Based on 998 bootstrap replicates
##
## CALL :
## boot::boot.ci(boot.out = boo03, L = inf_val)
##
## Intervals :
## Level Normal Basic Studentized
## 95% ( 0.1044, 0.4326 ) ( 0.0890, 0.4067 ) ( 0.0822, 0.4385 )
##
## Level Percentile BCa
## 95% ( 0.1503, 0.4680 ) ( 0.1408, 0.4466 )
## Calculations and Intervals on Original ScaleBootstrap CI With Transformation
Given that \(R^2\) is bounded, it may be more accurate to first transform the \(R^2\) estimates to an unbounded scale, obtain the CIs on the transformed scale, and then back transform it to between 0 and 1. This can be done in boot::boot.ci() as well with the logistic transformation:
boot::boot.ci(boo03, L = inf_val, h = qlogis,
# Need the derivative of the transformation
hdot = function(x) 1 / (x - x^2),
hinv = plogis)## BOOTSTRAP CONFIDENCE INTERVAL CALCULATIONS
## Based on 998 bootstrap replicates
##
## CALL :
## boot::boot.ci(boot.out = boo03, L = inf_val, h = qlogis, hdot = function(x) 1/(x -
## x^2), hinv = plogis)
##
## Intervals :
## Level Normal Basic Studentized
## 95% ( 0.1440, 0.4629 ) ( 0.1449, 0.4572 ) ( 0.1184, 0.4187 )
##
## Level Percentile BCa
## 95% ( 0.1503, 0.4680 ) ( 0.1408, 0.4466 )
## Calculations on Transformed Scale; Intervals on Original ScaleNote that the transformation only affects the Normal, Basic, and Studentized CIs.
Conclusion
This post demonstrates how to use multilevel bootstrapping to obtain CIs for \(R^2\). The post only focuses on marginal \(R^2\), but CIs for other \(R^2\) measures can be similarly obtained. The studentized CI is the most complex as it requires obtaining the sampling variance of \(R^2\) for each bootstrap sample. So far, to my knowledge, there has not been studies on which CI(s) perform best, so simulation studies are needed.
Further readings on multilevel bootstrap: